While our previous example focused on multi-class classification, where each image tile was assigned a single dominant label, many real-world biological scenarios require a more flexible approach. In histopathological images, for instance, it is common for tissue regions to contain both cancer cells and immune infiltrates, such as lymphocytes. These cell types are not mutually exclusive and frequently co-exist within the same field of view. In such cases, a classification system that only permits one label per image would be insufficient, as it would force the model to disregard relevant co-occurring structures.
This is where multi-label classification (MLC) becomes essential. Unlike multi-class classification, which restricts the model to returning a single category, MLC allows an image to be assigned multiple labels simultaneously. The model evaluates each possible class independently, calculating the likelihood of its presence or absence without forcing a binary choice between categories. Each class is treated as its own prediction task, allowing the model to output a vector of probabilities across all classes, with more than one potentially surpassing the decision threshold. This enables the model to return combinations of labels that more accurately reflect the complexity of the image content.
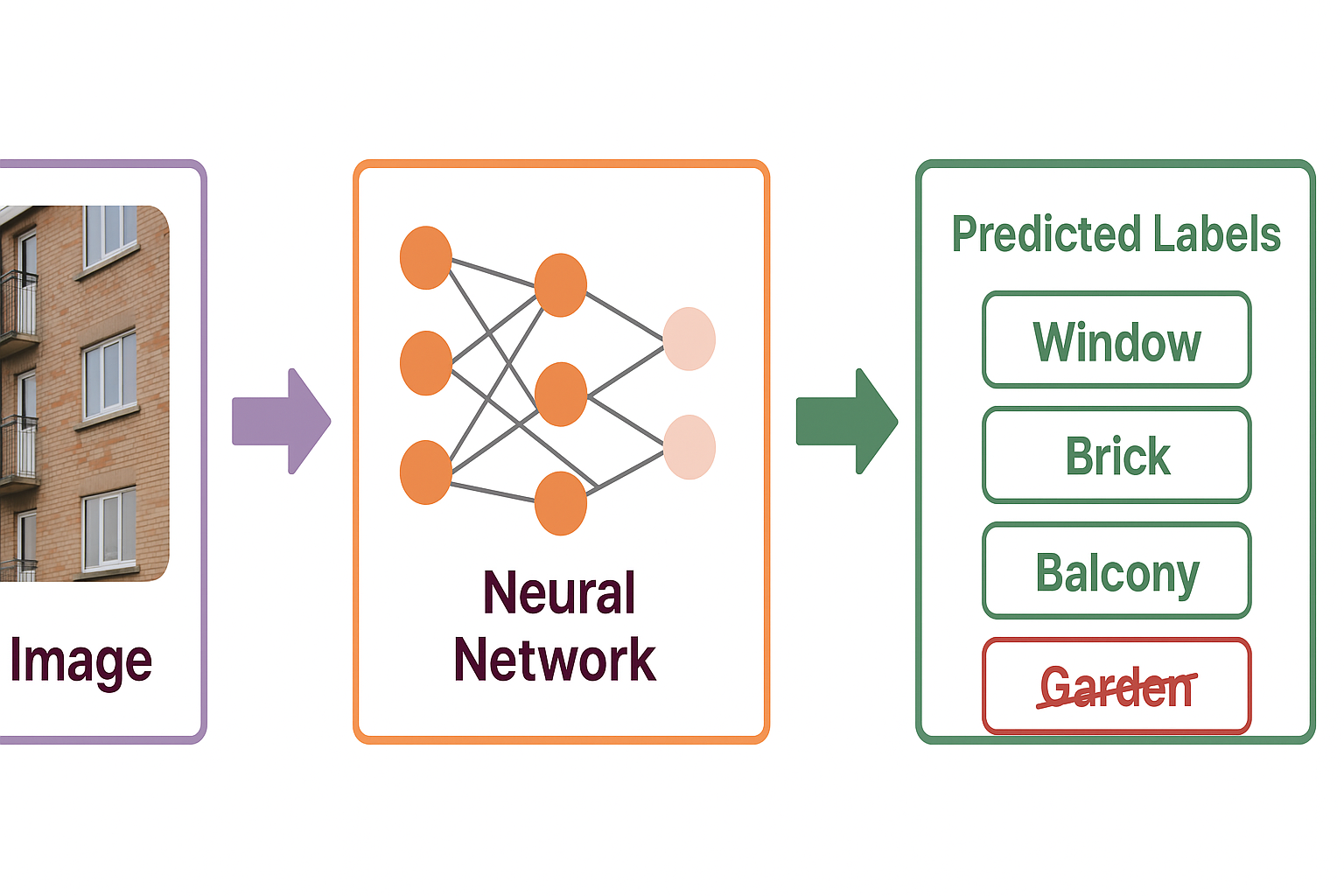
Multi-label classification workflow: An input image of a building is processed through a neural network to predict multiple labels (Window, Brick, Balcony) that can co-exist, unlike single-class prediction
MLC is particularly useful in biological and environmental research, where overlapping features and heterogeneous compositions are common. Whether identifying multiple cell populations within a tissue sample, detecting species cohabiting in an ecosystem or annotating overlapping structures in imaging datasets, multi-label models offer a richer and more realistic interpretation of image data. By capturing co-occurrence rather than enforcing artificial separation, they support more nuanced insights, improve the utility of automated annotation, and reduce information loss in complex visual scenes.
Multi-Label Classification Tutorial Video
The tutorial video below summarises the workflow and examples used in the multi-label classification section of our Custom Vision resource. Please consider its use as a supplement to the written materials, which give more depth and context.
Fruit Bowl Example
To illustrate the concept of multi-label classification, we'll switch to a more intuitive example: a dataset of AI-generated images showing different combinations of fruit in a wicker basket. Just as histological tiles might contain both cancer cells and lymphocytes, these images may feature more than one type of fruit.

Example fruit basket image containing multiple items (apple, bananas, basket) that would require multiple labels in a multi-label classification system
The labels or tags for this dataset should include:
- banana
- red_apple
- green_apple
- orange
- basket
Some images may include only one of these features; others may contain two or more. For this example, we will be uploading a total of 10 training images, and manually tagging them with the necessary labels, based on our visual interpretation.
The terms 'tag' and 'label' here are essentially referring to the same thing in this context. Custom Vision terms labels that are manually identified and added by the user as 'tags'.